Research
Complex molecular networks control cell type-specific gene expression. Dissecting the underlying principles is critical for understanding the cause of human diseases and for developing treatments. However, such investigation remains highly challenging in cancer due to the stochastic accumulation of genetic and epigenetic alternations in the heterogeneous cellular populations. Technology advancements in single-cell genomics provided excellent opportunities to access the comprehensive profiles from complex tissues. We focused on both development of cutting-edge genomic technologies for multimodal single-cell analysis and applying these technologies to study the combined effects of multiple regulatory layers in cell fate specification and maintenance during development, aging and diseases.
Genomics technologies
We will employ multidisciplinary approaches to develop innovative multimodal single-cell genomics tools for simultaneous analysis of the inputs (genetic variation or external stimuli), outputs (transcriptome) together with multiple regulatory layers from the same cells to dissect how the complex molecular networks function in cell fate specification and maintenance.
Functional impacts of DNA damages
DNA damages very frequently occur due to various factors including exogenous stimuli and endogenous metabolites. The stochastic nature of DNA damage events will produce cell populations with distinct stress responses. Canonical analysis to connect DNA damages with functional outputs was obscured by this inherent heterogeneity. We will develop and utilize novel multimodal single-cell genomics tools to study the impacts of DNA damage on epigenome integrity in aging, diseases, and cancer.
Molecular analysis of gene regulation
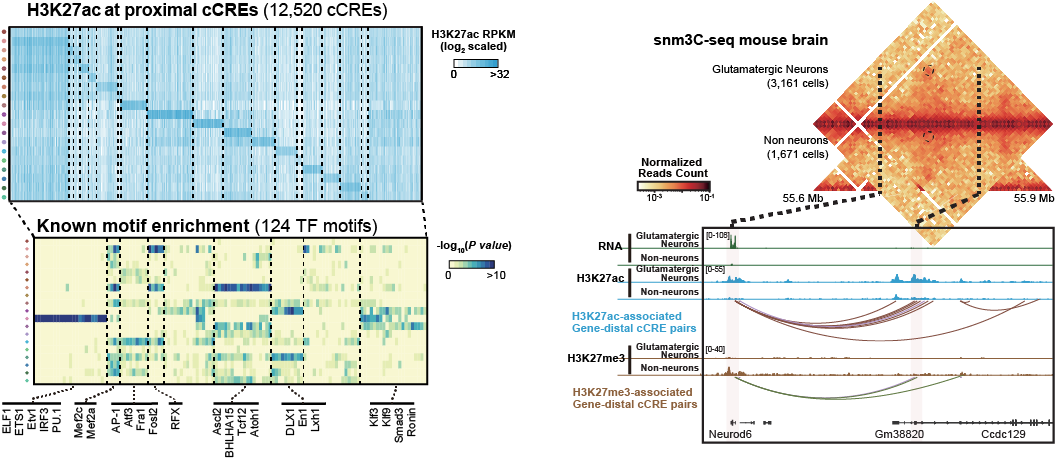
Extensive crosstalk between different regulatory layers has essential functions in controlling cell self-renewal and differentiation. With cutting-edge genomic technologies, we will carry out integrated molecular analysis from single cells to reconstruct the cell types & lineages resolved epigenome maps during mammalian development and diseases and to dissect the crosstalk and underlying regulatory mechanisms of multiple molecular modalities during these dynamic processes.
Technologies
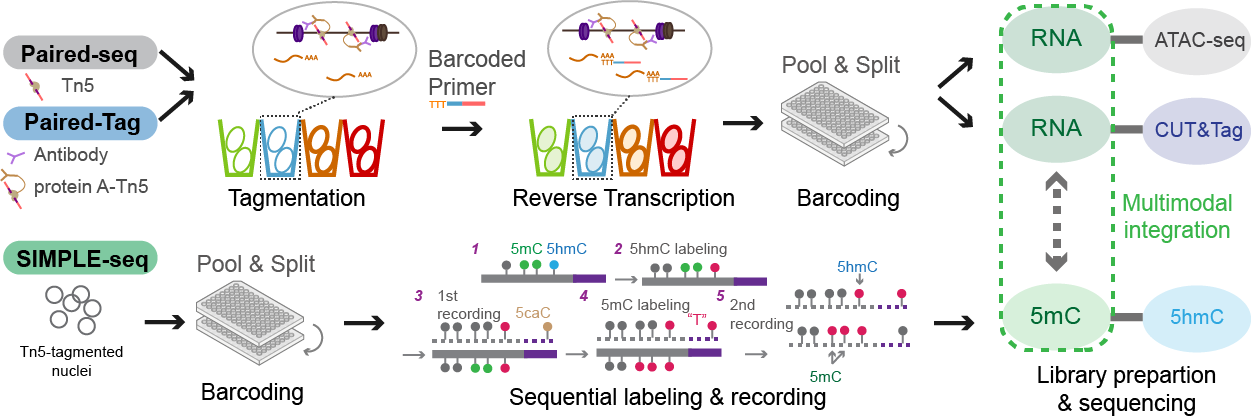
Paired-Damage-seq
Paired-Damage-seq can simultaneously profile oxidative & single-stranded DNA damage with gene expression from single cells. With this tool, we revealed the distribution of DNA damage hotspots exhibiting cell-type specific patterns.
SIMPLE-seq
SIMPLE-seq can map 5mC and 5hmC at single-cell & single-molecule resolution. Integrated analysis of these two epigenetic modifications reveals distinct epigenetic patterns associated with divergent regulatory programs in different cell states.
Paired-Tag/Droplet Paired-Tag
Paired-Tag and droplet Paired-Tag are methods for joint profiling of histone modifications and transcriptome in single cells to produce cell-type-resolved maps of chromatin state and transcriptome in complex tissues.
Paired-seq
Paired-seq is an ultra-high-throughput method for simultaneous analysis of gene expression and chromatin accessibility in millions of single cells.
CLEVER-seq
Based on chemical labelling, CLEVER-seq is a bisulfite-free method to detects 5fC, a 5mC-derived epigenetic modification, at single-cell and single-base resolution.
For more information, please visit our publications page.