Analyzing DNA damage, together with transcriptome, from single cells.
The faithful preservation of the genome template by DNA damage repair machinery and the precise control of gene expression patterns by epigenetic programs are essential for cells’ proper functioning in multicellular organisms. However, errors can arise during both processes and accumulate over time, resulting in the loss of molecular fidelity. Emerging evidence suggests the crosstalk between the maintenance of the genome and the epigenome stabilities, particularly in the contexts of cancer and aging. Whether such DNA damage-associated gene regulation program changes could long-term persist and lead to pathologic phenotypes in a programmed manner remained less clear. To elucidate the underlying mechanisms governing this process, it is essential to robustly measure DNA damage with gene expression programs from complex biological systems; however, such analysis has been hindered by the absence of proper methodologies.
Inspired by DNA base excision repair, we designed an in situ “labeling-by-repair” approach to convert the DNA damage signal to biotin. To confirm the specificity and sensitivity of this labeling approach, we first performed in vitro validation experiments and comparisons with relevant public datasets3. This approach is compatible with our previously developed Paired-Tag method for capturing of targeted genome regions jointly with gene expression analyses. By combining them, we developed the Paired-Damage-seq to parallelly analyze gene expression with oxidative and single-stranded DNA damage from single cells. To demonstrate the utilities of Paired-Damage-seq in studying the influences of DNA damage on gene regulation, as the proof-of-concept, we applied Paired-Damage-seq to both cultured HeLa cells and the heterogeneous mouse brain tissue.
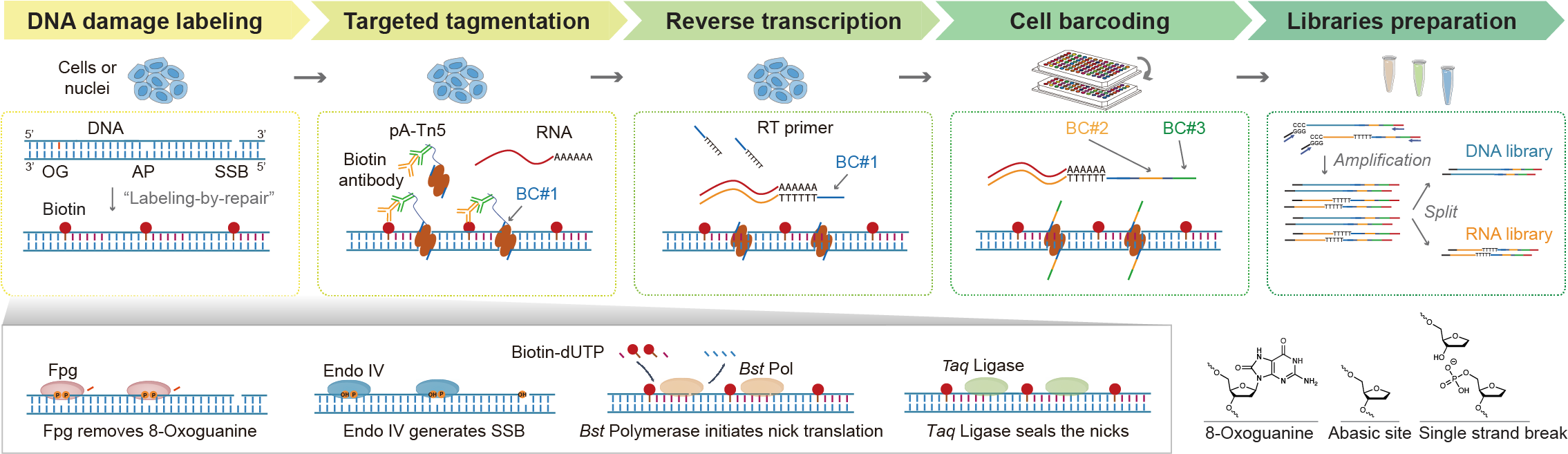
Consistent with prior studies, we observed the distribution of DNA damage hotspots is not randomly distributed but determined by both the base sequence contents and the local epigenetic states. By introducing oxidative stress to the cultured cells, we observed the extent of epigenetic information loss correlated with the accumulation of inducible oxidative DNA damage. Concurrently, gene expression level changes detected in parallel were linked to DNA damage induced at regulatory elements. By analyzing mouse brain tissue, we further observed the DNA damage hotspots, exhibiting cell-type-specific patterns, coincided with regions with age-associated epigenome decay. Furthermore, this selective genome vulnerability may exacerbate the risk of pathological gene programs through the cumulative effects of DNA damage and epigenome erosion over time.
DNA damage is a well-known hallmark of aging, while how different types of DNA damage contribute to specific molecular program changes during aging remained less clear. Paired-Damage-seq links DNA damage accumulation to their potential functional consequences, at a genome-wide scale, which opens new opportunities to identify targets of potential clinical interventions.
Read the paper, and visit the GitHub Page.
Back