Paired-Tag, now in droplets!
We report Droplet Paired-Tag, a rapid and robust method to simultaneously profile histone modifications and gene expression in single cells at scale. Droplet Paired-Tag provides researchers with a tool for studying the epigenome and gene regulation in complex tissues and disease pathogenesis.
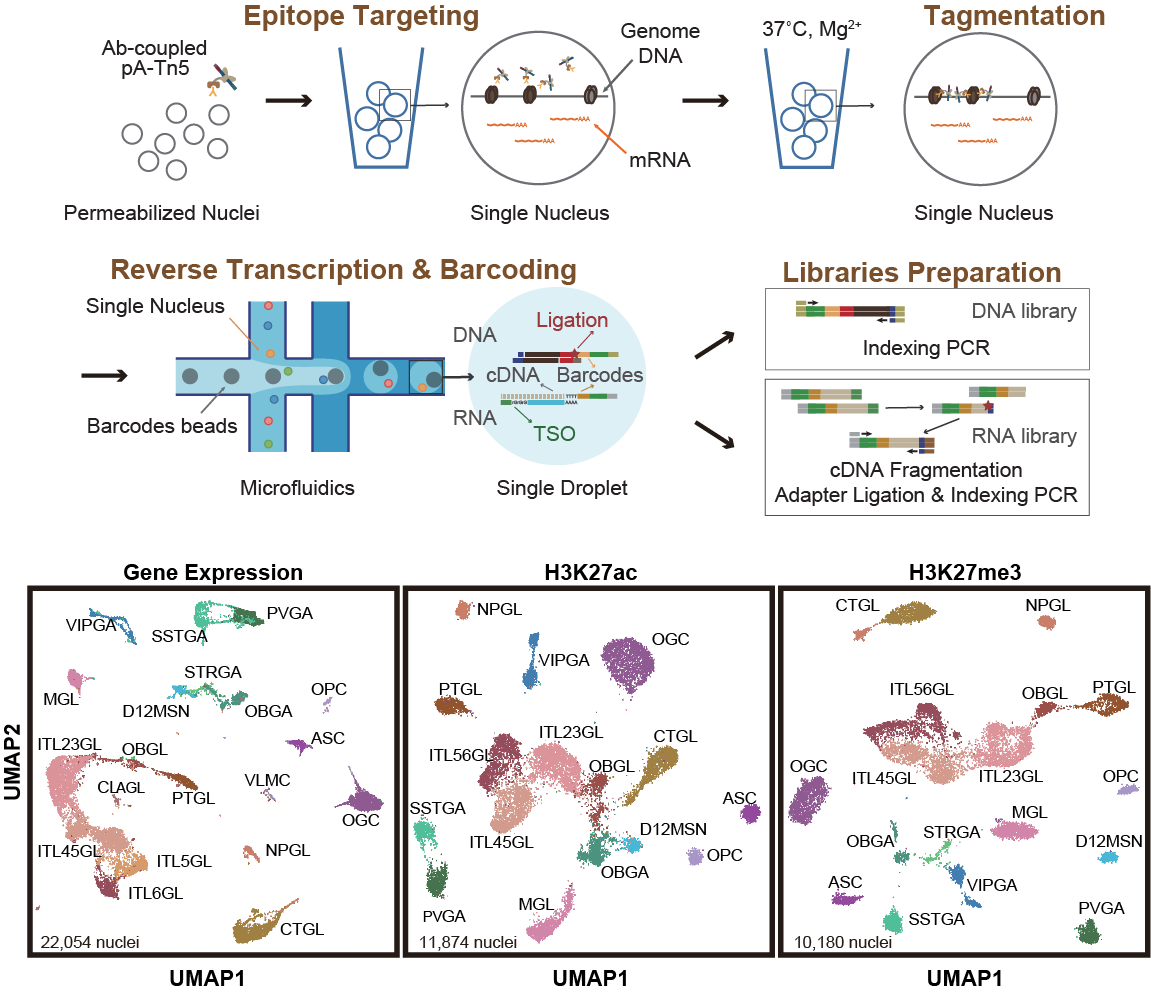
We developed Droplet Paired-Tag, a method which builds upon our combinatorial indexing-based Paired-Tag protocol but adapts a commercially available microfluidics platform (10x Chromium Single Cell Multiome ATAC + Gene Expression) to introduce cellular barcodes into droplets (upper panel). This approach results in a much-simplified protocol for jointly assaying histone marks and the transcriptome. We modified the existing CUT&Tag method by pre-coupling the protein A-Tn5 transposase fusion proteins (pA-Tn5) with primary antibodies against specific histone modifications. To make our pre-coupled pA-Tn5 fusion protein compatible with the commercially available platform, we designed the pA-Tn5 adaptor with a 3'-extended bridge sequence that is reverse complementary to the capturing oligo on the barcoded beads. We benchmarked the performance of Droplet Paired-Tag against several existing approaches using mouse embryonic stem cells and adult mouse frontal cortex.
These improvements mean that Droplet Paired-Tag — from the preparation of nuclei to the construction of the sequencing library — can be performed in less than 1.5 days, which is considerably shorter than the 3 days needed for the existing Paired-Tag procedure. In addition, Droplet Paired-Tag generates datasets with improved sensitivity, and the data also improved the prediction of putative candidate CRE–gene pairs by capturing stronger linkages between candidate CREs and genes over background than the existing combinatorial indexing Paired-Tag method.
One application of Droplet Paired-Tag is to develop cell-type-resolved maps of histone modifications in the human brain, heart, liver, and other organs, in both normal and disease states, producing a comprehensive cell census in complex tissues. The results would enable better classification of common and rare cell types in complex tissues, identification of the transcriptional regulatory elements, and characterization of their roles in development and disease pathogenesis.
Read the paper, and download the protocol.